16
Disease Prediction Based On Medical Diagnosis
In this article, we will discuss one of DOCTOR-Y's Machine Learning Models. This model predicts the current patients' medical conditions based on the previous diagnoses from the patient's medical history.
We used a dataset containing the diseases and their diagnosis and classified it using 3 different machine learning classifiers.
If you don't know what is DOCTOR-Y check this post.
Physicians will spend a lot of time reviewing the patient's previous e-prescriptions provided on DOCTOR-Y to know their past medical conditions and previous diseases.
That's why DOCTOR-Y provides a summarized chart representing the percentages for suffering from a group of diseases based on previous diagnoses. The model is provided with a dataset to train and classify these diseases. The model takes the diagnoses as input from previous prescriptions, and the output will be the predicted disease based on these diagnoses.
The snippet below shows how the model works.
python NLP.py "The patient has high blood pressure"
->['Hypertension']In this model, most of the data were collected from Disease Symptom Prediction Dataset from Kaggle.
Our dataset is used for the disease diagnosis model based on previous diagnoses, and it is divided into two columns the disease name, and diagnoses for that disease. We have 773 rows with 41 unique diseases leaving us with approximately 19 entries for each disease.
Our dataset is used for the disease diagnosis model based on previous diagnoses, and it is divided into two columns the disease name, and diagnoses for that disease. We have 773 rows with 41 unique diseases leaving us with approximately 19 entries for each disease.
The dataset is balanced. However, we faced a problem regarding building it from scratch. This data may lead to misclassification for diseases based on different diagnoses, which will affect the model’s accuracy.
The majority of the data is collected by hand from multiple healthcare sites; we looked carefully for definitions and diagnoses for the required diseases and ensured that no entries were duplicated.
| Prognosis | Prognosis | Prognosis | Prognosis |
|---|---|---|---|
| Fungal infection | Migraine | hepatitis A | Heart attack |
| Allergy | Cervical spondylosis | Hepatitis B | Varicose veins |
| GERD | Paralysis(brain hemorrhage) | Hepatitis C | Hypothyroidism |
| Chronic cholestasis | Jaundice | Hepatitis D | Hyperthyroidism |
| Drug Reaction | Malaria | Hepatitis E | Hypoglycemia |
| Peptic ulcer diseae | Chicken pox | Alcoholic hepatitis | Osteoarthristis |
| AIDS | Dengue | Tuberculosis | Arthritis |
| Diabetes | Typhoid | Common Cold | (vertigo) Paroymsal Positional Vertigo |
| Gastroenteritis | Psoriasis | Pneumonia | Acne |
| Bronchial Asthma | Impetigo | Dimorphic hemmorhoids(piles) | Urinary tract infection |
| Hypertension |
We prepared the data to be cleaner to obtain better results, and we implemented the following preprocessors:
We used three classification algorithms to process this data which are :
| Algorithm | Accuracy |
|---|---|
| SVM | 81% |
| NLP (LSTM) | 69.20% |
| NAÏVE BAYES | 74% |
The accuracy of the NLP model in training nearly reached 90% accuracy in training and 69.2% accuracy in the validation phase.

The least performing model was the LSTM model, while the best performing model was the SVM and Naiive model
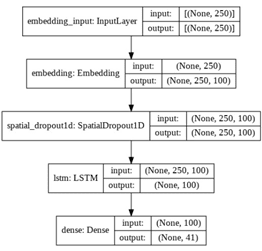
Unfortunately, papers did not provide guidelines on configuring the network of this model. So we had to use trial and error to choose the hyperparameters.
The results of the LSTM model are worse than both the SVM and Naïve models by achieving 69% accuracy; because the LSTM model reads the data sequentially and it has a memory that helps to keep words and use them in the prediction process, so it is more reliable than both.
We used the Diseases Diagnoses Prediction Model's results and combined them with the Diseases Symptoms Prediction Model's results to calculate the percentage of suffering from a group of diseases based on previous diagnoses + the associated symptoms.
The final diseases and their percentages are sent to the system server, which sends them to the client-side to be represented on a chart as shown in the figure below.

16